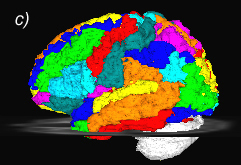
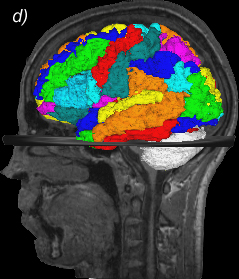
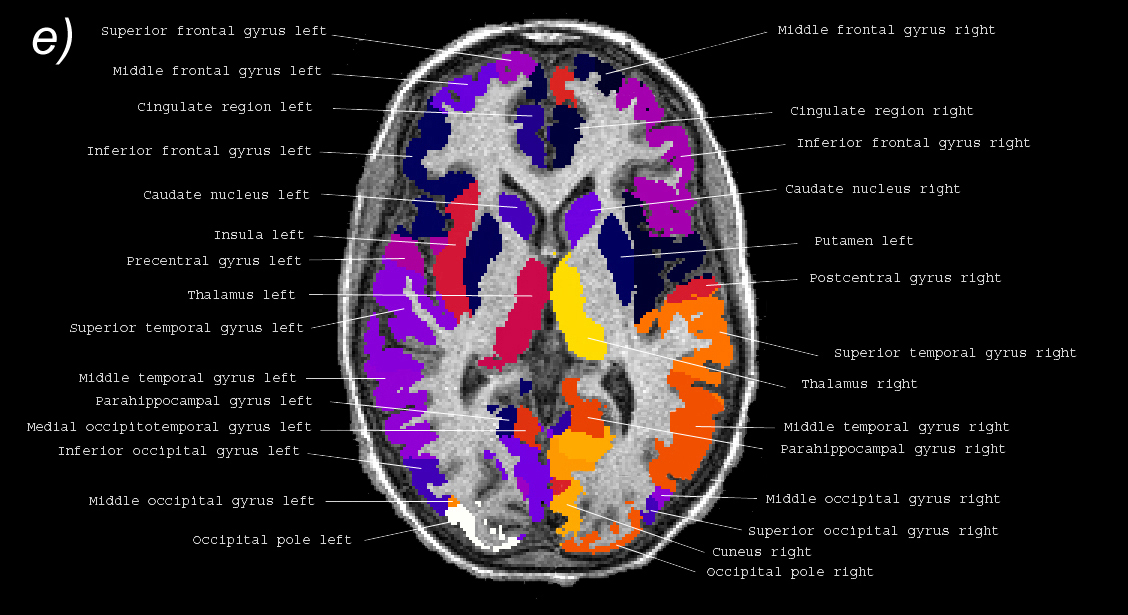
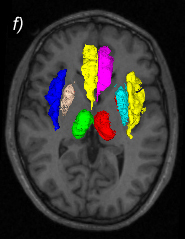
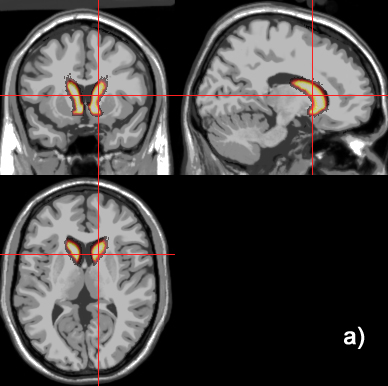
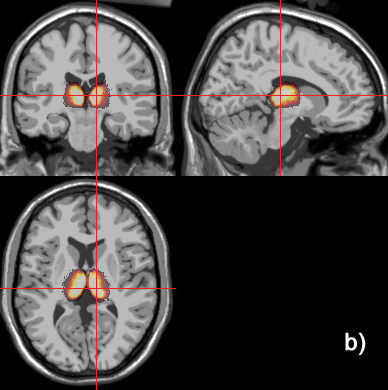
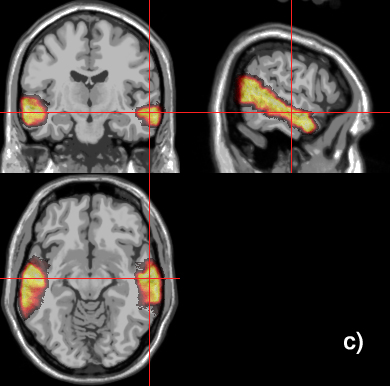
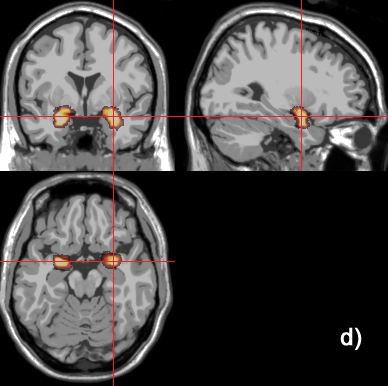
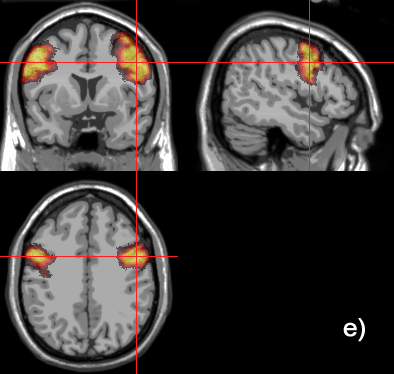
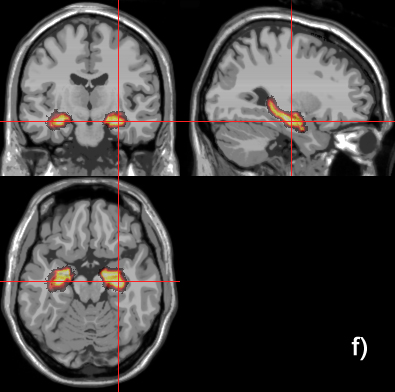
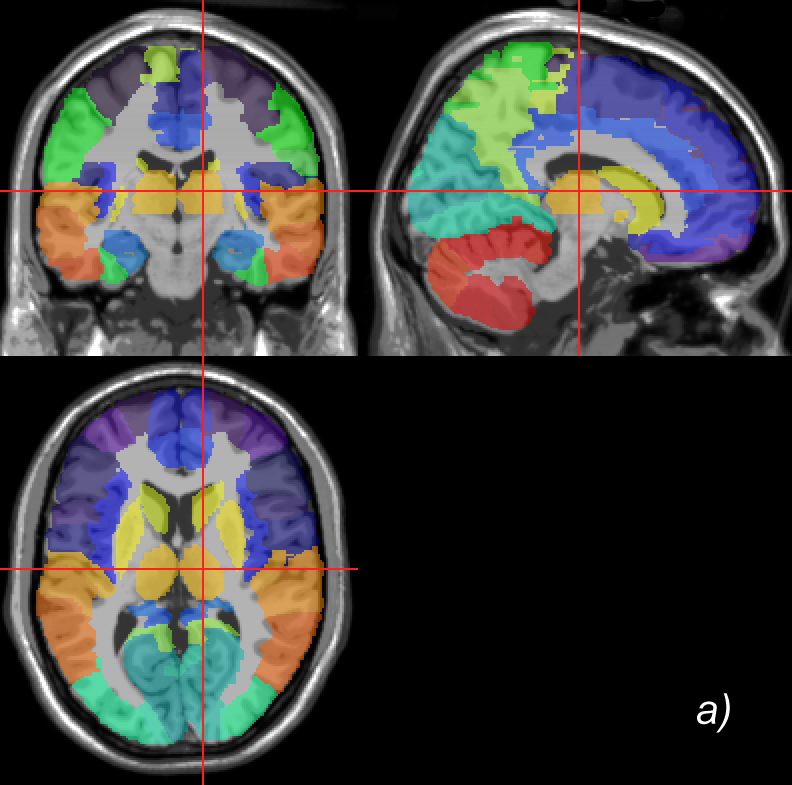
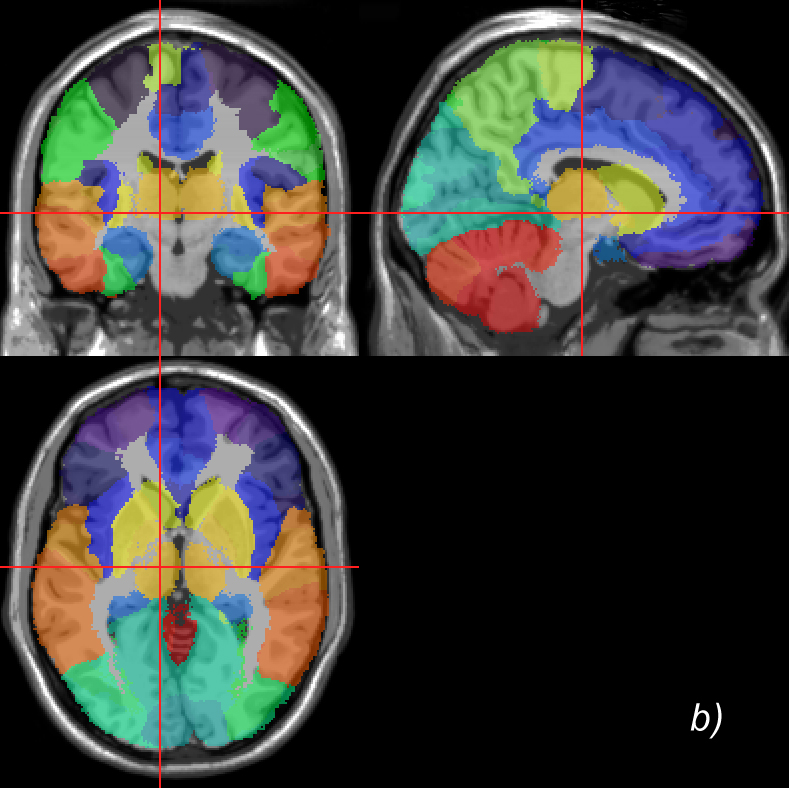
Related references:
- Collins D L, Holmes C, Peters T, Evans A. Automatic 3D Model-Based Neuroanatomical Segmentation. Human Brain Mapping. Vol.3: 190-208.1995.
- Collins D. , A. Zijdenbos, and A. Evans. Automatic Volume Estimation of Gross Cerebral Structures. 4th International Conference on Functional Mapping of the Human Brain, Montreal, June 1998. Organization for Human Brain Mapping. abstract no. 702.
- Collins D. L., A. P. Zijdenbos, W. F. C. Barré, and A. C. Evans, ``ANIMAL+INSECT: Inproved cortical structure segmentation,'' in Proc. of the Annual Symposium on Information Processing in Medical Imaging (A. Kuba, M. Samal, and A. Todd-Pokropek, eds.), vol. 1613 of LNCS, pp. 210-223, Springer, 1999.
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain.Neuron. 2002 Jan 31;33. (3):341-55.
- Fischl B, Van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V,Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004 Jan; 14(1):11-22.
- Maldjian JA,Laurienti PJ, Kraft RA, Burdette JH. An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage. 2003 Jul;19(3):1233-9.
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N. Automated anatomical labelling of activations in spm using a macroscopic anatomical parcellation of the MNI MRI single subject brain. Neuroimage 15: 273-289. 2002.
- Mazziotta J. , Toga A. , Evans A. , Fox P. and Lancaster. J. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping. NeuroImage, 2(2):89-101, 1995.
|
Disclaimer:
ALL INFORMATION, SOFTWARE, DOCUMENTATION, AND PUBLICATIONS ON THIS SITE ARE PROVIDED TO YOU "AS IS", WITHOUT WARRANTY OF ANY KIND AND WE HEREBY DISCLAIMS ALL WARRANTIES, EITHER EXPRESSED OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE IMPLIED WARRANTIES OF ACCURACY OR FITNESS FOR A PARTICULAR PURPOSE. |
|