Identifying microstates on the level of the individual EEG (first level clustering)
Before you start the microstate analysis, make sure that your data satisfies the following criteria
- Average reference
- No more significant artifacts
- Suitably filtered (the low-cut is typically at 1-2Hz, the hig-cut 20-30Hz)
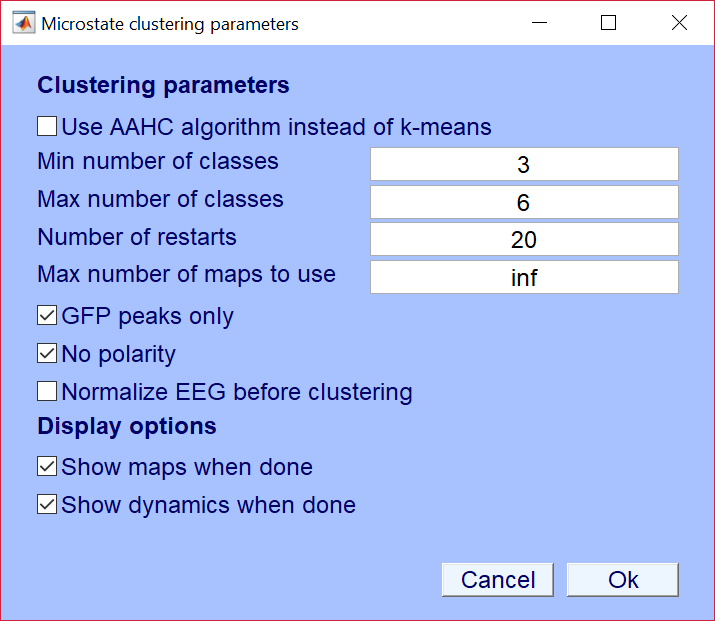
The identification of microstates on the level of the individual EEG is accessed thru the menu Tools->Microstates->Identify Microstates". This opens the following dialog:
Alternatively, the analysis step can also be started on the command line or using a script. In this case, the function to be called is
[EEG,com] = pop_FindMSTemplates(EEG,ClustPar,ShowMaps,ShowDyn);
typically followed by
[ALLEEG, EEG, CURRENTSET] = eeg_store(ALLEEG, EEG, CURRENTSET);
where the EEG contains the structure with the data to be clustered, EEGOUT is the data structure with the obtained clusters, com is a string with the command necessary to replicate the computation, ShowMaps and ShowDyn are boolean variables to toggle on or off the display of the results, and ClustPar is a structure that corresponds to the parameters of the above dialog, containing the following:
- "Min number of classes" / ClustPar.MinClasses -> Minimal number of clusters to search for.
- "Max number of classes" / ClustPar.MaxClasses -> Maximum number of clusters to search for.
- "Number of restarts" / ClustPar.Restarts -> Number of times the k-means is restarted with a new random configuration (I use about 20 to 50).
- "Max number of maps to use" / ClustPar.MaxMaps -> Use a random subsample of the data to identify the clusters (make this inf to include all data).
- "GFP peaks only" / ClustPar.GFPPeaks -> Limit the selection of maps used for cluster to moments of GFP peaks (often the standard).
- "No polarity" / ClustPar.IgnorePolarity -> Assign maps with inverted polarity to the same class (standard for resting EEG).
- "Use AAHC Algorithm" / ClustPar.UseAAHC -> Use the AAHC algorithm instead of the k-means (I prefer the k-means).
- "Normalize EEG before clustering" / ClustPar.UseAAHC -> Make all data GFP = 1 before clustering (I prefer not to).
For those wanting access to the result in matlab directly, this creates a sub-structure in the EEG called msinfo that is pretty self-explaining.